November 8, 2019 | Jola Glotzer
Structure of chromatin remodeling complex SWI/SNF resolved
Two CBC Awards to Han Yan and Yuan He, NU, have supported research recently published a preprint in bioRxiv, describing the structure of SWI/SNF complex bound to a nucleosome at near atomic resolution
Postdoctoral fellow Han Yan (left) and his mentor Yuan He, NU, are first and senior authors on the publication
Chromatin can either be in an active state (euchromatin; allowing gene expression) or inactive state (heterochromatin; gene expression is silenced). The transition between these two states requires dynamic remodeling of chromatin architecture to allow (or block) access of the transcription machinery to the condensed genomic DNA. In general, chromatin remodeling can be achieved by two mechanisms: covalent histone modifications by specific enzymes, (e.g., histone acetyltransferases, deacetylases, methyltransferases, and kinases) or by interacting with ATP-dependent complexes capable of moving or even ejecting nucleosomes. Aberrations in chromatin remodeling processes have been associated with human diseases, including cancer. In fact, targeting chromatin remodeling has become a “hot” therapeutic strategy in cancer treatment.
A recent study published in bioRxiv addresses the latter mechanism of chromatin remodeling. Specifically, using cryo-EM, the authors analyze the structure of an ATP-dependent multi-subunit chromatin remodeling complex SWI/SNF, which is involved in regulating the accessibility of DNA during transcription and gene repair. Looking at different configurations of the complex in ATP/ADP bound states, the authors propose a compelling model of how SWI/SNF complex recognizes and remodels nucleosomes resulting in “eviction” of the histone octamers, generating nucleosome-depleted chromatin regions and thus activating gene expression.
Two CBC Awards are acknowledged as having provided funds to support the research reported in this publication: a CBC Postdoctoral Research (PDR) Award (2016) and a CBC Catalyst Award (2015). The PDR Award was given to Han Yan — a postdoctoral fellow in Yuan He’s group at NU, and first author on the bioRxiv paper. Yuan He is senior author on the study and recipient of the Catalyst Award. He is Assistant Professor in the Department of Molecular Biosciences at NU.
Congratulations to all authors involved in the published work!
Publication attributed to *CBC funding:
Han Y, Reyes AA, Malik S, He Y. Cryo-electron microscopy structure of a nucleosome-bound SWI/SNF chromatin remodeling complex. bioRxiv preprint first posted online Oct. 16, 2019. (biorxiv.org)
ABSTRACT:
The multi-subunit chromatin remodeling complex SWI/SNF is highly conserved from yeast to humans and plays critical roles in various cellular processes including transcription and DNA damage repair. It uses the energy from ATP hydrolysis to remodel chromatin structure by sliding and evicting the histone octamer, creating DNA regions that become accessible to other essential protein complexes. However, our mechanistic understanding of the chromatin remodeling activity is largely hindered by the lack of a high-resolution structure of any complex from this family. Here we report the first structure of SWI/SNF from the yeast S. cerevisiae bound to a nucleosome at near atomic resolution determined by cryo-electron microscopy (cryo-EM). In the structure, the Arp module is sandwiched between the ATPase and the Body module of the complex, with the Snf2 HSA domain connecting all modules. The HSA domain also extends into the Body and anchors at the opposite side of the complex. The Body contains an assembly scaffold composed of conserved subunits Snf12 (SMARCD/BAF60), Snf5 (SMARCB1/BAF47/ INI1) and an asymmetric dimer of Swi3 (SMARCC/BAF155/170). Another conserved subunit Swi1 (ARID1/BAF250) folds into an Armadillo (ARM) repeat domain that resides in the core of the SWI/SNF Body, acting as a molecular hub. In addition to the interaction between Snf2 and the nucleosome, we also observed interactions between the conserved Snf5 subunit and the histones at the acidic patch, which could serve as an anchor point during active DNA translocation. Our structure allows us to map and rationalize a subset of cancer-related mutations in the human SWI/SNF complex and propose a model of how SWI/SNF recognizes and remodels the +1 nucleosome to generate nucleosome-depleted regions during gene activation.
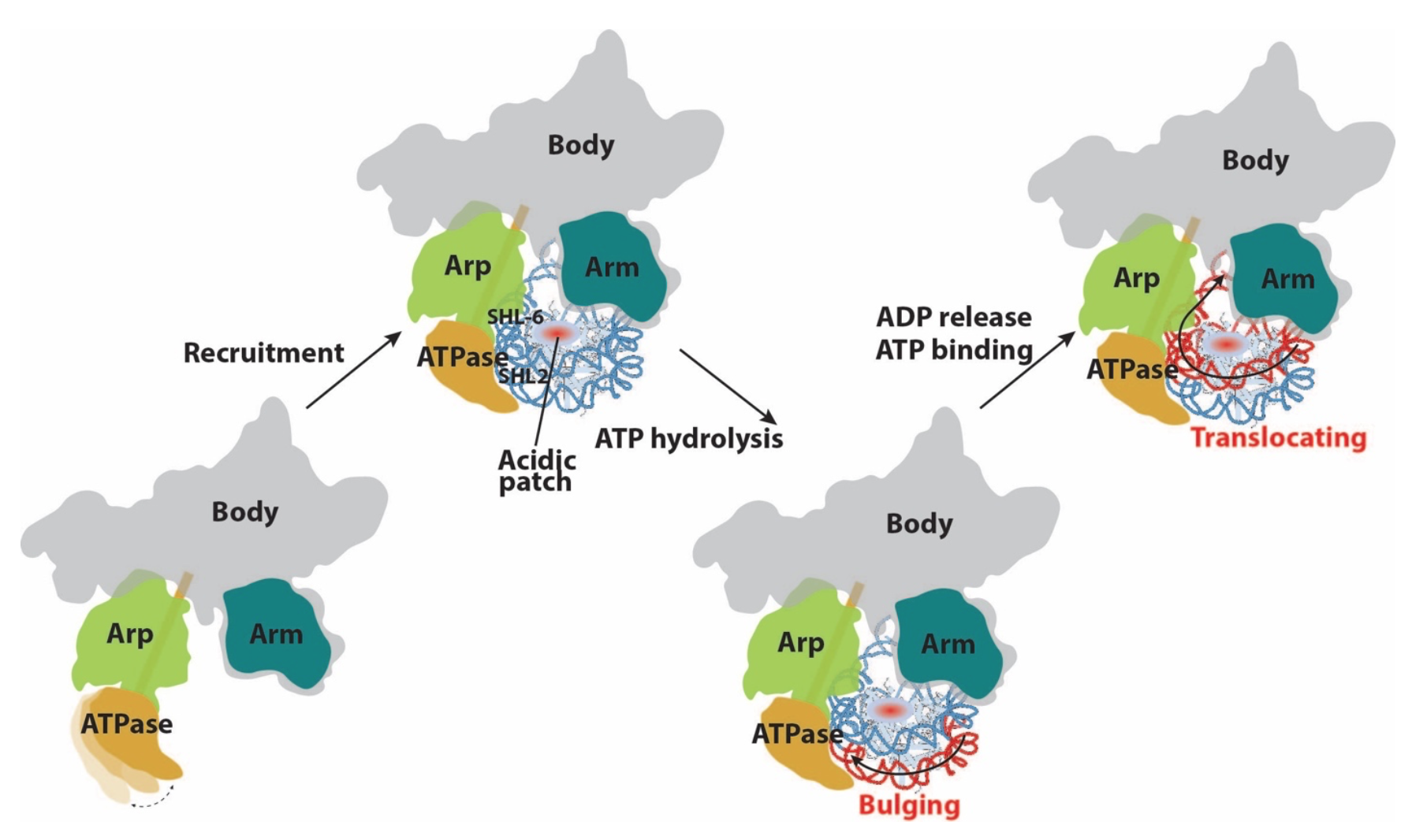
Model for chromatin remodeling activity by the SWI/SNF complex. Prior to engagement with nucleosome, the ATPase module of SWI/SNF is flexible. Nucleosome binding involves the ATPase and the HSA of Snf2 recognizing SHL2 and SHL-6, respectively. The Arm of SWI/SNF interact with the nucleosome surface near the acidic patch, likely serving as an anchor during active remodeling to keep the octamer from moving. Upon ATP hydrolysis, a bulge in the nucleosomal DNA is introduced at SHL 21, which is then propagated to the Exit side of the nucleosome when ADP is released and the next ATP molecule is bound, resulting in nucleosomal DNA translocation. (Source: bioRxiv)
ACKNOWLEDGMENTS:
We thank Dr. Jonathan Remis for assistance with microscope operation and data collection and Jason Pattie for computer support. We are grateful to Amy Rosenzweig, Ishwar Radhakrishnan and Susan Fishbain for helpful discussion and comments on the manuscript. We also thank the staff at the Structural Biology Facility (SBF) of Northwestern University for technical support. This work was supported by a Cornew Innovation Award from the Chemistry of Life Processes Institute at Northwestern University (to Y He), a Catalyst Award by the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust (to Y He), an Institutional Research Grant from the American Cancer Society (IRG-15-173-21 to Y He), an H Foundation Core Facility Pilot Project Award (to Y He). Y He is supported by P01 CA092584 and U54CA193419 from NIH/NCI. Y Han is a recipient of the Chicago Biomedical Consortium Postdoctoral Research Grant.
Featured CBC Community member(s):
Yan Han and Yuan He, NU
- *CBC Postdoctoral Research Award (2016):
▸ Proteomic and Structural Analysis of Human RNA Pol III Transcription Machinery
PIs: Yan Han (postdoc) and Yuan He (NU) - *CBC Catalyst Award (2015):
▸ Toward the Visualization of Spliceosomal Intermediates
PIs: Yuan He (NU), Joseph Piccirilli and Jonathan Staley (UChicago)
